Joerg Hiller
Jul 25, 2024 02:15
NVIDIA introduces VISTA-2D, a model enhancing cell segmentation and morphology clustering in spatial omics, offering a pivotal advancement for biological research.
NVIDIA has unveiled VISTA-2D, a foundational model designed to significantly improve cell segmentation in cell imaging and spatial omics workflows, according to NVIDIA Technical Blog. This model aims to enhance the accuracy of downstream tasks by leveraging advanced image embedding techniques.
Feature Extraction and Clustering
The VISTA-2D model employs an image encoder to generate embeddings that can be transformed into segmentation masks. These embeddings provide essential information about cell morphologies, allowing for precise cell segmentation. NVIDIA’s blog post explains that these embeddings can be clustered to group cells with similar morphologies automatically.
To demonstrate the model’s capabilities, NVIDIA has provided a detailed Jupyter notebook that walks users through the process of segmenting cells and extracting their spatial features using VISTA-2D. The notebook also shows how to cluster these features using RAPIDS, creating an automated pipeline for classifying cell types.
Prerequisites and Setup
Users interested in exploring the VISTA-2D model need a basic understanding of Python, Jupyter, and Docker. The Docker container required for this tutorial can be initiated with the following command:
docker run --rm -it \
-v /path/to/this/repo/:/workspace \
-p 8888:8888 \
--gpus all \
nvcr.io/nvidia/pytorch:24.03-py3 \
/bin/bash
Additional Python packages needed for the tutorial can be installed using:
pip install -r requirements.txt
Cell Segmentation with VISTA-2D
The initial step involves loading a VISTA-2D model checkpoint and using it to segment cells in an image. The segmentation process generates a feature vector for each cell, which contains all necessary information for cell morphology analysis. These vectors are then used in clustering algorithms to group cells with similar features.
Segmenting Cells
The segmentation function processes the cell image through VISTA-2D, resulting in segmentation masks that label each cell individually. This allows for accurate feature extraction for each cell.
img_path="example_livecell_image.tif" patch, segmentation, pred_mask = segment_cells(img_path, model_ckpt)
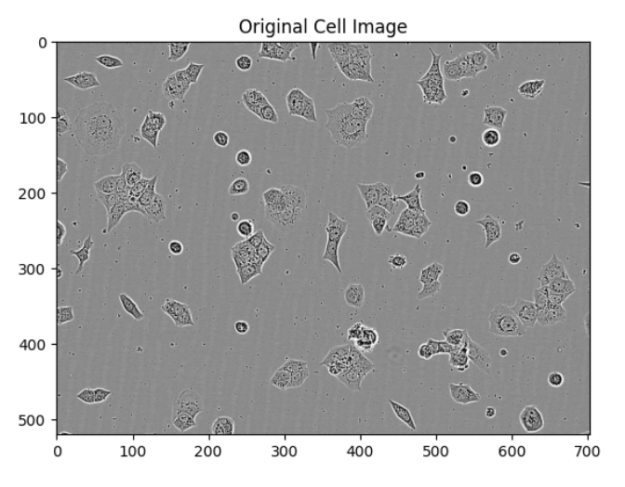
Plotting Segmentation
The segmented images can be visually verified using the plot_segmentation function. This function displays the original image, the segmentation result, and individual masks for each cell.
plot_segmentation(patch, segmentation, pred_mask)


Clustering Features with RAPIDS
Once feature vectors are extracted, they are clustered using RAPIDS, a GPU-accelerated machine learning library. The TruncatedSVD algorithm reduces the dimensionality of the feature vectors, making it easier to visualize clusters in 3D space.
dim_red_model = TruncatedSVD(n_components=3) X = dim_red_model.fit_transform(cell_features)
The DBSCAN algorithm is then used to cluster the reduced feature vectors. This method assigns cluster labels to each cell, which can be visualized using Plotly for an interactive 3D plot.
model = DBSCAN(eps=0.003, min_samples=2) labels = model.fit_predict(X)

Conclusion
NVIDIA’s VISTA-2D model offers a significant advancement in cell imaging and spatial omics by providing accurate cell segmentation and feature extraction. Coupled with RAPIDS for clustering, this model enables efficient classification of cell types, paving the way for more detailed and automated biological research.
Image source: Shutterstock
Credit: Source link






















 Bitcoin
Bitcoin  Ethereum
Ethereum  XRP
XRP  Tether
Tether  Solana
Solana  USDC
USDC  Dogecoin
Dogecoin  Cardano
Cardano  Lido Staked Ether
Lido Staked Ether  TRON
TRON  Wrapped Bitcoin
Wrapped Bitcoin  Chainlink
Chainlink  Wrapped stETH
Wrapped stETH  Avalanche
Avalanche  Sui
Sui  Stellar
Stellar  Litecoin
Litecoin  Toncoin
Toncoin  Shiba Inu
Shiba Inu  Hedera
Hedera  LEO Token
LEO Token  USDS
USDS  Hyperliquid
Hyperliquid  Polkadot
Polkadot  WETH
WETH  MANTRA
MANTRA  Bitcoin Cash
Bitcoin Cash  Bitget Token
Bitget Token  Ethena USDe
Ethena USDe  Wrapped eETH
Wrapped eETH  Uniswap
Uniswap  Monero
Monero  NEAR Protocol
NEAR Protocol  Pepe
Pepe  WhiteBIT Coin
WhiteBIT Coin  Aave
Aave  Bittensor
Bittensor  Ondo
Ondo  Aptos
Aptos  Internet Computer
Internet Computer  Dai
Dai  Official Trump
Official Trump  Ethereum Classic
Ethereum Classic  Mantle
Mantle  Tokenize Xchange
Tokenize Xchange  OKB
OKB  Gate
Gate  sUSDS
sUSDS  Sonic (prev. FTM)
Sonic (prev. FTM) 
